libcbm run of tutorial 2 from CBM-CFS3¶
[1]:
import os, json
import pandas as pd
%matplotlib inline
Import the required packages from libcbm
- sit_cbm_factory: a module for initializing the CBM model from the CBM Standard import tool format
- cbm_simulator: simulates the sit dataset using the CBM model
- libcbm.resources: gets files for tutorial 2 that are bundled in libcbm
[9]:
from libcbm.input.sit import sit_cbm_factory
from libcbm.model.cbm import cbm_simulator
from libcbm.model.cbm.cbm_output import CBMOutput
from libcbm import resources
from libcbm.storage.backends import BackendType
Setup¶
Load the standard import tool configuration at the specified path. This configuration encompasses the data source for the various sit inputs (sit_inventory, sit_classifiers etc.) and also the relationships of classifiers, and disturbance types to the default CBM parameters.
[10]:
config_path = os.path.join(
resources.get_test_resources_dir(), "cbm3_tutorial2", "sit_config.json"
)
sit = sit_cbm_factory.load_sit(config_path)
Initialize and validate the inventory in the sit dataset
[11]:
classifiers, inventory = sit_cbm_factory.initialize_inventory(sit)
Create storage for CBM simulation results. This is a built in method, and it is not mandatory since it is possible to interact with the simulation dataframes directly as well.
[12]:
cbm_output = CBMOutput(
classifier_map=sit.classifier_value_names,
disturbance_type_map=sit.disturbance_name_map,
)
Simulation¶
The following line of code spins up the CBM inventory and runs it through 100 timesteps.
[13]:
with sit_cbm_factory.initialize_cbm(sit) as cbm:
# Create a function to apply rule based disturbance events and transition rules based on the SIT input
rule_based_processor = sit_cbm_factory.create_sit_rule_based_processor(
sit, cbm
)
# The following line of code spins up the CBM inventory and runs it through 200 timesteps.
cbm_simulator.simulate(
cbm,
n_steps=200,
classifiers=classifiers,
inventory=inventory,
pre_dynamics_func=rule_based_processor.pre_dynamics_func,
reporting_func=cbm_output.append_simulation_result,
backend_type=BackendType.numpy,
)
Pool Results¶
[17]:
pi = cbm_output.classifiers.to_pandas().merge(
cbm_output.pools.to_pandas(),
left_on=["identifier", "timestep"],
right_on=["identifier", "timestep"],
)
[18]:
pi.head()
[18]:
| identifier | timestep | Working_Species_Or_Leading_Species | Site_Quality | Density_Class | Working_Status | Input | SoftwoodMerch | SoftwoodFoliage | SoftwoodOther | ... | BelowGroundSlowSoil | SoftwoodStemSnag | SoftwoodBranchSnag | HardwoodStemSnag | HardwoodBranchSnag | CO2 | CH4 | CO | NO2 | Products | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | B | G | D | W | 200.0 | 0.000000 | 0.000000 | 0.0 | ... | 10529.242115 | 5568.605479 | 1544.809522 | 0.0 | 0.0 | 690032.809697 | 631.259013 | 5681.261733 | 0.0 | 0.0 |
| 1 | 2 | 0 | B | G | D | W | 100.0 | 0.003124 | 0.376897 | 0.0 | ... | 5273.531854 | 2666.157167 | 666.417417 | 0.0 | 0.0 | 345255.294728 | 315.629506 | 2840.630866 | 0.0 | 0.0 |
| 2 | 3 | 0 | B | G | D | W | 100.0 | 0.030782 | 1.867345 | 0.0 | ... | 5278.708266 | 2553.024910 | 574.973378 | 0.0 | 0.0 | 345471.445778 | 315.629506 | 2840.630866 | 0.0 | 0.0 |
| 3 | 4 | 0 | B | G | D | W | 100.0 | 0.117354 | 4.744316 | 0.0 | ... | 5281.125622 | 2444.693447 | 496.077048 | 0.0 | 0.0 | 345669.654559 | 315.629506 | 2840.630866 | 0.0 | 0.0 |
| 4 | 5 | 0 | B | G | D | W | 100.0 | 0.303283 | 9.168135 | 0.0 | ... | 5281.488695 | 2340.959458 | 428.006664 | 0.0 | 0.0 | 345853.464520 | 315.629506 | 2840.630866 | 0.0 | 0.0 |
5 rows × 33 columns
[19]:
biomass_pools = [
"SoftwoodMerch",
"SoftwoodFoliage",
"SoftwoodOther",
"SoftwoodCoarseRoots",
"SoftwoodFineRoots",
"HardwoodMerch",
"HardwoodFoliage",
"HardwoodOther",
"HardwoodCoarseRoots",
"HardwoodFineRoots",
]
dom_pools = [
"AboveGroundVeryFastSoil",
"BelowGroundVeryFastSoil",
"AboveGroundFastSoil",
"BelowGroundFastSoil",
"MediumSoil",
"AboveGroundSlowSoil",
"BelowGroundSlowSoil",
"SoftwoodStemSnag",
"SoftwoodBranchSnag",
"HardwoodStemSnag",
"HardwoodBranchSnag",
]
biomass_result = pi[["timestep"] + biomass_pools]
dom_result = pi[["timestep"] + dom_pools]
total_eco_result = pi[["timestep"] + biomass_pools + dom_pools]
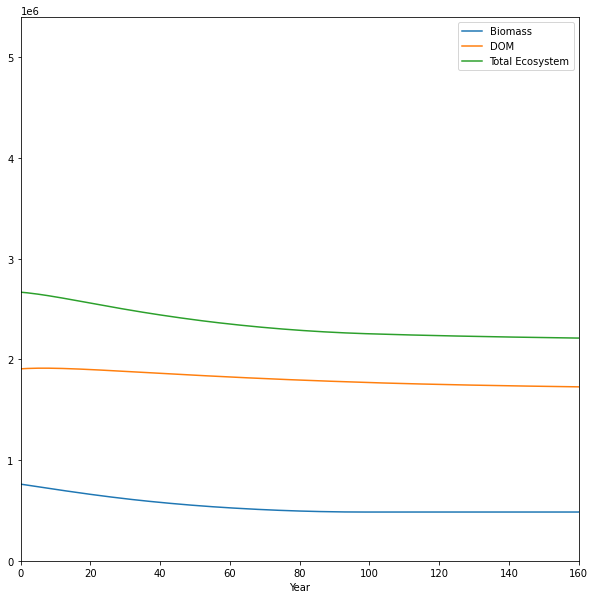
annual_carbon_stocks = pd.DataFrame(
{
"Year": pi["timestep"],
"Biomass": pi[biomass_pools].sum(axis=1),
"DOM": pi[dom_pools].sum(axis=1),
"Total Ecosystem": pi[biomass_pools + dom_pools].sum(axis=1),
}
)
annual_carbon_stocks.groupby("Year").sum().plot(
figsize=(10, 10), xlim=(0, 160), ylim=(0, 5.4e6)
)
[19]:
<AxesSubplot:xlabel='Year'>
State Variable Results¶
[23]:
si = cbm_output.state.to_pandas()
[24]:
si.head()
[24]:
| identifier | timestep | last_disturbance_type | time_since_last_disturbance | time_since_land_class_change | growth_enabled | enabled | land_class | age | growth_multiplier | regeneration_delay | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | N | 0 | -1 | 1 | 1 | 0 | 0 | 1.0 | 0 |
| 1 | 2 | 0 | N | 1 | -1 | 1 | 1 | 0 | 1 | 1.0 | 0 |
| 2 | 3 | 0 | N | 2 | -1 | 1 | 1 | 0 | 2 | 1.0 | 0 |
| 3 | 4 | 0 | N | 3 | -1 | 1 | 1 | 0 | 3 | 1.0 | 0 |
| 4 | 5 | 0 | N | 4 | -1 | 1 | 1 | 0 | 4 | 1.0 | 0 |
[25]:
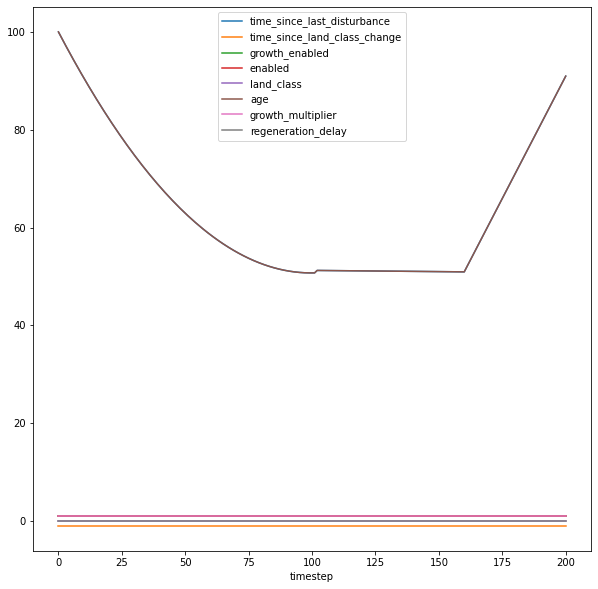
state_variables = [
"timestep",
"time_since_last_disturbance",
"time_since_land_class_change",
"growth_enabled",
"enabled",
"land_class",
"age",
"growth_multiplier",
"regeneration_delay",
]
si[state_variables].groupby("timestep").mean().plot(figsize=(10, 10))
[25]:
<AxesSubplot:xlabel='timestep'>
Flux Indicators¶
[30]:
fi = cbm_output.flux.to_pandas()
[31]:
fi.head()
[31]:
| identifier | timestep | DisturbanceCO2Production | DisturbanceCH4Production | DisturbanceCOProduction | DisturbanceBioCO2Emission | DisturbanceBioCH4Emission | DisturbanceBioCOEmission | DecayDOMCO2Emission | DisturbanceSoftProduction | ... | DisturbanceVFastBGToAir | DisturbanceFastAGToAir | DisturbanceFastBGToAir | DisturbanceMediumToAir | DisturbanceSlowAGToAir | DisturbanceSlowBGToAir | DisturbanceSWStemSnagToAir | DisturbanceSWBranchSnagToAir | DisturbanceHWStemSnagToAir | DisturbanceHWBranchSnagToAir | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 2 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 3 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 4 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 5 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
5 rows × 54 columns
[32]:
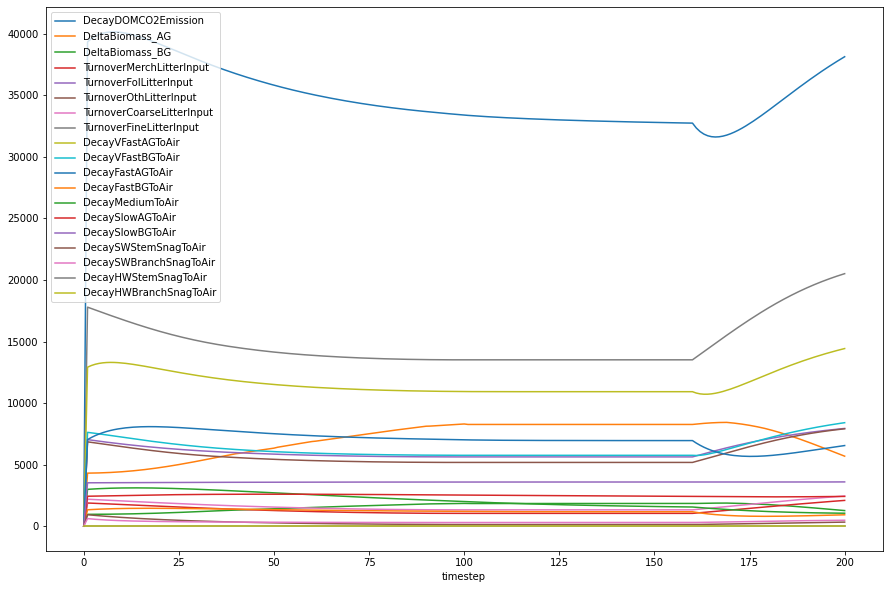
annual_process_fluxes = [
"DecayDOMCO2Emission",
"DeltaBiomass_AG",
"DeltaBiomass_BG",
"TurnoverMerchLitterInput",
"TurnoverFolLitterInput",
"TurnoverOthLitterInput",
"TurnoverCoarseLitterInput",
"TurnoverFineLitterInput",
"DecayVFastAGToAir",
"DecayVFastBGToAir",
"DecayFastAGToAir",
"DecayFastBGToAir",
"DecayMediumToAir",
"DecaySlowAGToAir",
"DecaySlowBGToAir",
"DecaySWStemSnagToAir",
"DecaySWBranchSnagToAir",
"DecayHWStemSnagToAir",
"DecayHWBranchSnagToAir",
]
[33]:
fi[["timestep"] + annual_process_fluxes].groupby("timestep").sum().plot(
figsize=(15, 10)
)
[33]:
<AxesSubplot:xlabel='timestep'>
Appendix¶
SIT source data¶
[34]:
sit.sit_data.age_classes
[34]:
| name | class_size | start_year | end_year | |
|---|---|---|---|---|
| 0 | AGEID0 | 0 | 0 | 0 |
| 1 | AGEID1 | 10 | 1 | 10 |
| 2 | AGEID2 | 10 | 11 | 20 |
| 3 | AGEID3 | 10 | 21 | 30 |
| 4 | AGEID4 | 10 | 31 | 40 |
| 5 | AGEID5 | 10 | 41 | 50 |
| 6 | AGEID6 | 10 | 51 | 60 |
| 7 | AGEID7 | 10 | 61 | 70 |
| 8 | AGEID8 | 10 | 71 | 80 |
| 9 | AGEID9 | 10 | 81 | 90 |
| 10 | AGEID10 | 10 | 91 | 100 |
| 11 | AGEID11 | 10 | 101 | 110 |
| 12 | AGEID12 | 10 | 111 | 120 |
| 13 | AGEID13 | 10 | 121 | 130 |
| 14 | AGEID14 | 10 | 131 | 140 |
| 15 | AGEID15 | 10 | 141 | 150 |
| 16 | AGEID16 | 10 | 151 | 160 |
| 17 | AGEID17 | 10 | 161 | 170 |
| 18 | AGEID18 | 10 | 171 | 180 |
| 19 | AGEID19 | 10 | 181 | 190 |
| 20 | AGEID20 | 10 | 191 | 200 |
[35]:
sit.sit_data.inventory
[35]:
| Working_Species_Or_Leading_Species | Site_Quality | Density_Class | Working_Status | age | area | delay | land_class | historical_disturbance_type | last_pass_disturbance_type | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BF | GOOD | D1 | W | 0 | 200 | 0 | 0 | DISTID1 | DISTID1 |
| 1 | BF | GOOD | D1 | W | 1 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 2 | BF | GOOD | D1 | W | 2 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 3 | BF | GOOD | D1 | W | 3 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 4 | BF | GOOD | D1 | W | 4 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 196 | BF | GOOD | D1 | W | 196 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 197 | BF | GOOD | D1 | W | 197 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 198 | BF | GOOD | D1 | W | 198 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 199 | BF | GOOD | D1 | W | 199 | 100 | 0 | 0 | DISTID1 | DISTID1 |
| 200 | BF | GOOD | D1 | W | 200 | 100 | 0 | 0 | DISTID1 | DISTID1 |
201 rows × 10 columns
[36]:
sit.sit_data.classifiers
[36]:
| id | name | |
|---|---|---|
| 0 | 1 | Working_Species_Or_Leading_Species |
| 1 | 2 | Site_Quality |
| 2 | 3 | Density_Class |
| 3 | 4 | Working_Status |
[37]:
sit.sit_data.classifier_values
[37]:
| classifier_id | name | description | |
|---|---|---|---|
| 1 | 1 | BF | Balsam Fir |
| 2 | 1 | BS | Black Spruce |
| 3 | 1 | JL | Western Larch |
| 4 | 1 | JP | Jack Pine |
| 5 | 1 | OS | Other Spruce |
| 6 | 1 | RP | Red Pine |
| 7 | 1 | SH | Unspecified Softwood |
| 8 | 1 | LT | Larch |
| 9 | 1 | WS | White Spruce |
| 11 | 2 | GOOD | Good |
| 12 | 2 | MEDIUM | Medium |
| 13 | 2 | POOR | Poor |
| 15 | 3 | D0 | No Crown Closure |
| 16 | 3 | D1 | 75% - > Crown Closure |
| 17 | 3 | D2 | 51% - 74% Crown Closure |
| 18 | 3 | D3 | 26% - 50% Crown Closure |
| 19 | 3 | NN | Null Value For The Non-forested Polyg |
| 21 | 4 | W | Working forest |
| 22 | 4 | R | Reserve Forest |
[38]:
sit.sit_data.disturbance_types
[38]:
| sit_disturbance_type_id | id | name | |
|---|---|---|---|
| 0 | 1 | DISTID1 | Natural forest fire |
| 1 | 2 | DISTID2 | Senescence |
| 2 | 3 | DISTID3 | Mountain pine beetle infestation |
| 3 | 4 | DISTID4 | ClearCut Harvesting |
[41]:
sit.sit_data.disturbance_events
[41]:
| Working_Species_Or_Leading_Species | Site_Quality | Density_Class | Working_Status | min_age | max_age | MinYearsSinceDist | MaxYearsSinceDist | LastDistTypeID | MinTotBiomassC | ... | MinSWMerchStemSnagC | MaxSWMerchStemSnagC | MinHWMerchStemSnagC | MaxHWMerchStemSnagC | efficiency | sort_type | target_type | target | disturbance_type | time_step | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 1 |
| 1 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 2 |
| 2 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 3 |
| 3 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 4 |
| 4 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 5 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 155 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 156 |
| 156 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 157 |
| 157 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 158 |
| 158 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 159 |
| 159 | BF | GOOD | D1 | W | 80 | 200 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | 1 | SORT_BY_SW_AGE | Area | 200 | DISTID4 | 160 |
160 rows × 33 columns
[39]:
sit.sit_data.yield_table
[39]:
| Working_Species_Or_Leading_Species | Site_Quality | Density_Class | Working_Status | leading_species | v0 | v1 | v2 | v3 | v4 | ... | v11 | v12 | v13 | v14 | v15 | v16 | v17 | v18 | v19 | v20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | BF | GOOD | D1 | W | 29 | 0 | 0 | 0 | 8 | 20 | ... | 112 | 113 | 113 | 113 | 113 | 113 | 113 | 113 | 113 | 113 |
1 rows × 26 columns
[40]:
print(json.dumps(sit.config, indent=4, sort_keys=True))
{
"import_config": {
"age_classes": {
"params": {
"path": "age_classes.csv"
},
"type": "csv"
},
"classifiers": {
"params": {
"path": "classifiers.csv"
},
"type": "csv"
},
"disturbance_types": {
"params": {
"path": "disturbance_types.csv"
},
"type": "csv"
},
"events": {
"params": {
"path": "disturbance_events.csv"
},
"type": "csv"
},
"inventory": {
"params": {
"path": "inventory.csv"
},
"type": "csv"
},
"transitions": {
"params": {
"path": "transition_rules.csv"
},
"type": "csv"
},
"yield": {
"params": {
"path": "growth_and_yield.csv"
},
"type": "csv"
}
},
"mapping_config": {
"disturbance_types": [
{
"default_dist_type": "Clearcut harvesting without salvage",
"user_dist_type": "ClearCut Harvesting"
},
{
"default_dist_type": "Wildfire",
"user_dist_type": "Natural forest fire"
},
{
"default_dist_type": "Stand\u2013replacing natural succession",
"user_dist_type": "Senescence"
},
{
"default_dist_type": "Insect disturbance",
"user_dist_type": "Mountain pine beetle infestation"
}
],
"nonforest": null,
"spatial_units": {
"admin_boundary": "Ontario",
"eco_boundary": "Boreal Shield East",
"mapping_mode": "SingleDefaultSpatialUnit"
},
"species": {
"species_classifier": "Working Species Or Leading Species",
"species_mapping": [
{
"default_species": "Balsam fir",
"user_species": "Balsam Fir"
},
{
"default_species": "Black spruce",
"user_species": "Black Spruce"
},
{
"default_species": "Western larch",
"user_species": "Western Larch"
},
{
"default_species": "Jack pine",
"user_species": "Jack Pine"
},
{
"default_species": "Other spruce",
"user_species": "Other Spruce"
},
{
"default_species": "Red pine",
"user_species": "Red Pine"
},
{
"default_species": "Unspecified softwood species",
"user_species": "Unspecified Softwood"
},
{
"default_species": "Tamarack/larch",
"user_species": "Larch"
},
{
"default_species": "White spruce",
"user_species": "White Spruce"
}
]
}
}
}
[ ]:
[ ]:
[ ]: